Experiences from constructing LLM-based (Mistral 7B) biomedical question-answering system on high of Qdrant and OpenSearch indices with hallucination detection technique
Final September (2023), we launched into the event of the VerifAI project, after receiving funding from the NGI Search funding scheme of Horizon Europe.
The concept of the challenge was to create a generative search engine for the biomedical area, based mostly on vetted paperwork (subsequently we used a repository of biomedical journal publications known as PubMed), with a further mannequin that might confirm the generated reply, by evaluating the referenced article and generated declare. In domains corresponding to biomedicine, but additionally usually, in sciences, there’s a low tolerance for hallucinations.
Whereas there are initiatives and merchandise, corresponding to Elicit or Perplexity, that do partially RAG (Retrieval-Augmented Technology) and may reply and reference paperwork for biomedical questions, there are a couple of elements that differentiate our challenge. Firstly, we focus for the time being on the biomedical paperwork. Secondly, as this can be a funded challenge by the EU, we decide to open-source the whole lot that we have now created, from supply code, fashions, mannequin adapters, datasets, the whole lot. Thirdly, no different product, that’s out there for the time being, does a posteriori verification of the generated reply, however they often simply depend on pretty easy RAG, which reduces hallucinations however doesn’t take away them fully. One of many primary goals of the challenge is to handle the difficulty of so-called hallucinations. Hallucinations in Massive Language Fashions (LLMs) confer with cases the place the mannequin generates textual content that’s plausible-sounding however factually incorrect, deceptive, or nonsensical. On this regard, the challenge provides to the reside system’s distinctive worth.
The challenge has been shared beneath the AGPLv3 license.
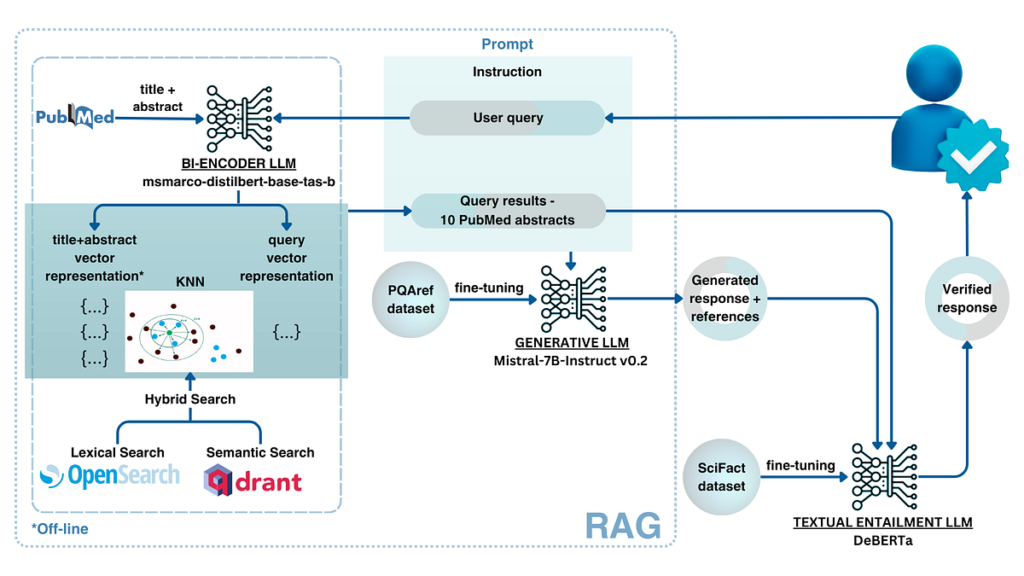
The general methodology that we have now utilized may be seen within the following determine.
When a person asks a query, the person question is remodeled into a question, and the data retrieval engine is requested for essentially the most related biomedical abstracts, listed in PubMed, for the given query. With a purpose to get hold of essentially the most related paperwork, we have now created each a lexical index, based mostly on OpenSearch, and a vector/semantic search based mostly on Qdrant. Specifically, lexical search is nice at retrieving related paperwork containing precise phrases because the question, whereas semantic search helps us to go looking semantic area and retrieve paperwork that imply the identical issues, however might phrase it in another way. The retrieved scores are normalized and a mixture of paperwork from these two indices are retrieved (hybrid search). The highest paperwork from the hybrid search, with questions, are handed into the context of a mannequin for reply technology. In our case, we have now fine-tuned the Mistral 7B-instruct mannequin, utilizing QLoRA, as a result of this allowed us to host a reasonably small well-performing mannequin on a reasonably low cost cloud occasion (containing NVidia Tesla T4 GPU with 16GB of GPU RAM). After the reply is generated, the reply is parsed into sentences and references that help these sentences and handed to the separate mannequin that checks whether or not the generated declare is supported by the content material of the referenced summary. This mannequin classifies claims into supported, no-evidence, and contradicting courses. Lastly, the reply, with highlighted claims that might not be absolutely supported by the abstracts, is introduced to the person.
Subsequently, the system has 3 elements — info retrieval, reply technology, and reply verification. Within the following sections, we are going to describe in additional element every of those sections.
From the beginning of the challenge, we geared toward constructing a hybrid search, combining semantic and lexical search. The preliminary concept was to create it utilizing a single software program, nevertheless, that turned out not really easy, particularly for the index of PubMed’s measurement. PubMed comprises about 35 million paperwork, nevertheless, not all comprise full abstracts. There are outdated paperwork, from the Nineteen Forties and Nineteen Fifties that will not have abstracts, however as properly some information paperwork and related which have solely titles. We now have listed solely the paperwork containing full abstracts and ended up with about 25 million paperwork. PubMed unpacked is about 120GB in measurement.
It isn’t problematic to create a well-performing index for lexical search in OpenSearch. This labored just about out of the field. We now have listed titles, and summary texts, and added some metadata for filtering, like yr of publication, authors, and journal. OpenSearch helps FAISS as a vector retailer. So we tried to index our knowledge with FAISS, however this was not doable, because the index was too massive and we had been working out of reminiscence (we had a 64GB cloud occasion for index). The indexing was performed utilizing MSMarco fine-tuned mannequin based mostly on DistilBERT (sentence-transformers/msmarco-distilbert-base-tas-b). Since we discovered that FAISS solely supported the in-memory index, we would have liked to seek out one other resolution that might be capable of retailer part of the index on the laborious drive. The answer was discovered within the Qdrant database, because it helps in-memory mapping and on-disk storage of part of the index.
One other concern that appeared whereas creating the index was that after we did reminiscence mapping and created the entire PubMed index, the question could be executed for a very long time (virtually 30 seconds). The issue was that calculations of dot merchandise in 32-bit precision had been taking some time for the index having 25 million paperwork (and doubtlessly loading elements of the index from HDD). Subsequently, we have now made a search utilizing solely 8-bit precision, and we have now diminished the time wanted from about 30 seconds to lower than a half second.
The lexical index contained entire abstracts, nevertheless, for the semantic index, paperwork wanted to be break up, as a result of the transformer mannequin we used for constructing the semantic index might swallow 512 tokens. Subsequently the paperwork had been break up on full cease earlier than the 512th token, and the identical occurred on each subsequent 512 tokens.
With a purpose to create a mixture of semantic and lexical search, we have now normalized the outputs of the queries, by dividing all scores returned both from Qdrant or OpenSearch by the highest rating returned for the given doc retailer. In that method, we have now obtained two numbers, one for semantic and the opposite for lexical search, within the vary between 0–1. Then we examined the precision of retrieved most related paperwork within the high retrieved paperwork utilizing the BioASQ dataset. The outcomes may be seen within the desk under.
We now have performed some re-ranking experiments utilizing full precision, so you possibly can see extra particulars within the paper. However this has not been used within the software on the finish. The general conclusion was that lexical search does a reasonably good job, and there may be some contribution of semantic search, with the very best efficiency obtained at weights of 0.7 for lexical search, and 0.3 for semantic.
Lastly, we have now constructed a question processing, the place for lexical querying stopwords had been excluded from the question, and search was carried out within the lexical index, and similarity was calculated for the semantic index. Values for paperwork from each semantic and lexical indexes had been normalized and summed up, and the highest 10 paperwork had been retrieved.
As soon as the highest 10 paperwork had been retrieved, we might cross these paperwork to a generative mannequin for referenced reply technology. We now have examined a number of fashions. This may be performed properly with GPT4-Turbo fashions, and most of commercially out there platforms would use GPT4 or Claude fashions. Nevertheless, we wished to create an open-source variant, the place we don’t rely on industrial fashions, having smaller and extra environment friendly fashions whereas having additionally efficiency near the efficiency of business fashions. Subsequently, we have now examined issues with Mistral 7B instruct in each zero-shot regime and fine-tuned utilizing 4bit QLora.
With a purpose to fine-tune Mistral, we would have liked to create a dataset for referenced query answering utilizing PubMed. We created a dataset by randomly deciding on questions from the PubMedQA dataset, then retrieved the highest 10 related paperwork and used GPT-4 Turbo for referenced reply technology. We now have known as this dataset the PQAref dataset and published it on HuggingFace. Every pattern on this dataset comprises a query, a set of 10 paperwork, and a generated reply with referenced paperwork (based mostly on 10 handed in context).
Utilizing this dataset, we have now created a QLoRA adapter for Mistral-7B-instruct. This was skilled on the Serbian National AI platform within the Nationwide Information Heart of Serbia, utilizing Nvidia A100 GPU. The coaching lasted round 32 hours.
We now have carried out an analysis evaluating Mistral 7B instruct v1, and Mistral 7B instruct v2, with and with out QLoRA fine-tuning (so with out is zero-shot, based mostly solely on instruction, whereas with QLoRA we might avoid wasting tokens as instruction was not essential for the immediate, as fine-tuning would make mannequin do what is required), and we in contrast it with GPT-4 Turbo (with immediate: “Reply the query utilizing related abstracts offered, as much as 300 phrases. Reference the statements with the offered abstract_id in brackets subsequent to the assertion.”). A number of evaluations concerning the variety of referenced paperwork and whether or not the referenced paperwork are related have been performed. These outcomes may be seen within the tables under.
From this desk, it may be concluded that Mistral, particularly the primary model in zero-shot (0-M1), does not likely usually reference context paperwork, regardless that it was requested within the immediate. However, the second model confirmed significantly better efficiency, nevertheless it was removed from GPT4-Turbo or fine-tuned Mistrals 7B. Positive-tuned Mistrals tended to quote extra paperwork, even when the reply might be present in one of many paperwork, and added some extra info in comparison with GPT4-Turbo.
As may be seen from the second desk, GPT4-Turbo missed related references solely as soon as in the entire check set, whereas Mistral 7B-instruct v2 with fine-tuning missed a bit extra, however confirmed nonetheless comparable efficiency, given a lot smaller mannequin measurement.
We now have additionally checked out a number of solutions manually, to verify the solutions make sense. In the long run, within the app, we’re utilizing Mistral 7B instruct v2 with a fine-tuned QLoRA adapter.
The ultimate a part of the system is reply verification. For reply verification, we have now developed a number of options, nevertheless, the primary one is a system that verifies whether or not the generated declare is predicated on the summary that was referenced. We now have fine-tuned a number of fashions on the SciFact dataset from the Allen Institute for AI with a number of BERT and Roberta-based fashions.
With a purpose to mannequin enter, we have now parsed the reply to seek out sentences and associated references. We now have discovered that the primary and final sentence in our system are sometimes introduction or conclusion sentences, and might not be referenced. All different sentences ought to have a reference. If a sentence contained a reference, it was based mostly on that PubMed doc. If the sentence doesn’t comprise a reference, however the sentence earlier than and after are referenced, we calculate the dot product of the embeddings of that sentence and sentences in 2 abstracts. Whichever summary comprises the sentence with the best dot product is handled because the summary that the sentence was based mostly on.
As soon as the reply is parsed and we have now discovered all of the abstracts the claims are based mostly on, we cross it to the fine-tuned mannequin. The enter to the mannequin was engineered within the following method
For deBERT-a fashions:
[CLS]declare[SEP]proof[SEP]
For Roberta-based fashions:
<s>declare</s></s>proof</s>
The place the declare is a generated declare from the generative part, and proof is the textual content of concatenated title and summary from referenced the PubMed doc.
We now have evaluated the efficiency of our fine-tuned fashions and obtained the next outcomes:
Typically, when the fashions are fine-tuned on the identical dataset as they’re examined, the outcomes are good. Subsequently, we wished to check it additionally on out-of-domain knowledge. So we have now chosen the HealthVer dataset, which can be within the healthcare area used for declare verification. The outcomes had been the next:
We additionally evaluated the SciFact label prediction process utilizing the GPT-4 mannequin (with a immediate “Critically asses whether or not the assertion is supported, contradicted or there isn’t a proof for the assertion within the given summary. Output SUPPORT if the assertion is supported by the summary. Output CONTRADICT if the assertion is in contradiction with the summary and output NO EVIDENCE if there isn’t a proof for the assertion within the summary.” ), leading to a precision of 0.81, a recall of 0.80, and an F-1 rating of 0.79. Subsequently, our mannequin has higher efficiency and as a result of a lot decrease variety of parameters was extra environment friendly.
On high of the verification utilizing this mannequin, we have now additionally calculated the closest sentence from the summary utilizing dot product similarity (utilizing the identical MSMarco mannequin we use for semantic search). We visualize the closest sentence to the generated one on the person interface by hovering over a sentence.
We now have developed a person interface, to which customers can register, log in, and ask questions, the place they are going to get referenced solutions, hyperlinks to PubMed, and verification by described posterior mannequin. Listed here are a couple of screenshots of the person interface:
We’re right here to current our expertise from constructing the VerifAI challenge, which is the biomedical generative question-answering engine with verifiable solutions. We’re open-sourcing entire code and fashions and we’re opening the applying, at the very least quickly (is determined by the finances for a way lengthy, and the way and whether or not we will discover a sustainable resolution for internet hosting). Within the subsequent part, you will discover the hyperlinks to the applying, web site, code, and fashions.
The applying is a results of the work of a number of individuals (see beneath Workforce part) and virtually a yr of analysis and improvement. We’re joyful and proud to current it now to a wider public and hope that individuals will take pleasure in it, and as properly contribute to it, to make it extra sustainable and higher sooner or later.
In case you use any of the methodology, fashions, datasets, or are mentioning this challenge in your paper’s background part, please cite a number of the following papers:
- Košprdić, M., Ljajić, A., Bašaragin, B., Medvecki, D., & Milošević, N. “Verif. ai: Towards an Open-Source Scientific Generative Question-Answering System with Referenced and Verifiable Answers.” The Sixteenth International Conference on Evolving Internet INTERNET 2024 (2024).
- Bojana Bašaragin, Adela Ljajić, Darija Medvecki, Lorenzo Cassano, Miloš Košprdić, Nikola Milošević “How do you know that? Teaching Generative Language Models to Reference Answers to Biomedical Questions”, The 23rd BioNLP Workshop 2024, Colocated with ACL 2024
The applying may be accessed and tried at https://verifai-project.com/ or https://app.verifai-project.com/. Customers can right here register, and ask inquiries to strive our platform.
The code of the challenge is out there at GitHub at https://github.com/nikolamilosevic86/verif.ai. The QLoRA adapters for Mistra 7B instruct may be discovered on HuggingFace at https://huggingface.co/BojanaBas/Mistral-7B-Instruct-v0.2-pqa-10 and https://huggingface.co/BojanaBas/Mistral-7B-Instruct-v0.1-pqa-10. A generated dataset for fine-tuning the generative part may be discovered additionally on HuggingFace at https://huggingface.co/datasets/BojanaBas/PQAref. A mannequin for reply verification may be discovered at https://huggingface.co/MilosKosRad/TextualEntailment_DeBERTa_preprocessedSciFACT.
The VerifAI challenge was developed as a collaborative challenge between Bayer Pharma R&D and the Institute for Synthetic Intelligence Analysis and Improvement of Serbia, funded by the NGI Search challenge beneath grant settlement No 101069364. The individuals concerned are Nikola Milosevic, Lorenzo Cassano, Bojana Bašaragin, Miloš Košprdić, Adela Ljajić and Darija Medvecki.